GlioBLAST: Establishing Prognosis and Targeted Therapy for Glioblastoma by Applying Convolutional Neural Networks to Detect Histological Features, Molecular Subtypes, MGMT Methylation, and EGFR Amplification from Brain-Biopsy Whole-Slide Images
Abstract:
Glioblastoma is the most aggressive and deadly malignant brain tumor, having a five-year survival rate of only 7% and accounting for 48% of all primary malignant brain tumors. The poor prognosis associated with most glioblastoma diagnoses results from heterogeneity between and within tumors. However, early detection and classification can significantly improve patient prognosis. Diagnosis and prognosis extensively rely on identifying tumor features (like pseudopalisading necrosis) and determining genetic biomarkers (like MGMT gene methylation and EGFR gene amplification) from biopsy imagery. Yet, manual image assessment requires days for diagnosis and encounters variability between pathologists. Artificial intelligence innovations in healthcare, like deep learning and computer vision, can address these constraints with accurate medical image analysis. Artificial intelligence has been applied to modalities like CT scans and MRI images, but few studies explore histopathological image analysis, even though histopathological diagnosis is the gold-standard classification method. I developed GlioBLAST, a novel neural-network system to identify tumor features and genetic biomarkers from histopathological whole-slide images (WSIs) of brain-biopsy tissue. With UNet and convolutional neural networks, applying transfer learning and fine-tuning, my models achieved 98% accuracy in classifying molecular subtypes, 94% accuracy in detecting MGMT methylation, and 97% accuracy in detecting EGFR amplification. I also developed a web application where medical professionals can upload WSIs and obtain the corresponding heatmaps of predictions for tumor features, molecular subtypes, and genetic factors. The application can perform tumor segmentation of a WSI in under 20 minutes and create molecular subtypes, MGMT gene methylation, and EGFR gene amplification heatmaps in under 10 minutes. The automated GlioBLAST system provides reliable and accurate glioblastoma detection to predict prognosis and potentially save lives.Bibliography/Citations:
“Astrocytoma - Overview - Mayo Clinic.” https://www.mayoclinic.org/diseases-conditions/astrocytoma/cdc-20350132 (accessed Jan. 02, 2022).
“Glioblastoma Multiforme.” https://www.aans.org/en/Patients/Neurosurgical-Conditions-and-Treatments/Glioblastoma-Multiforme (accessed Jan. 02, 2022).
“Gliomas.” https://www.hopkinsmedicine.org/health/conditions-and-diseases/gliomas (accessed Jan. 02, 2022).
“Glioblastoma - Overview - Mayo Clinic.” https://www.mayoclinic.org/diseases-conditions/glioblastoma/cdc-20350148 (accessed Jan. 02, 2022).
“Glioblastoma—Unraveling the Threads to Make Progress,” Aug. 03, 2017. https://www.cancer.gov/news-events/cancer-currents-blog/2017/glioblastoma-research-making-progress (accessed Jan. 02, 2022).
E. Calabrese, J. E. Villanueva-Meyer, and S. Cha, “A fully automated artificial intelligence method for non-invasive, imaging-based identification of genetic alterations in glioblastomas,” Scientific Reports, vol. 10, no. 1. 2020. doi: 10.1038/s41598-020-68857-8.
Allen Institute for Brain Science, “Ivy GAP Technical White Paper,” Allen Institute for Brain Science, May 2015.
“Glioma.” https://www.mayoclinic.org/diseases-conditions/glioma/diagnosis-treatment/drc-20350255 (accessed Jan. 02, 2022).
D. J. Pisapia, R. Magge, and R. Ramakrishna, “Improved Pathologic Diagnosis—Forecasting the Future in Glioblastoma,” Front. Neurol., vol. 8, p. 707, 2017.
“Glioblastoma - Overview - mayo clinic.” https://www.mayoclinic.org/diseases-conditions/glioblastoma/cdc-20350148 (accessed Jan. 02, 2022).
L. Zhao, V. H. F. Lee, M. K. Ng, H. Yan, and M. F. Bijlsma, “Molecular subtyping of cancer: current status and moving toward clinical applications,” Brief. Bioinform., vol. 20, no. 2, pp. 572–584, Mar. 2019.
R. G. W. Verhaak et al., “Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1,” Cancer Cell, vol. 17, no. 1, pp. 98–110, Jan. 2010.
P. Sidaway, “CNS cancer: Glioblastoma subtypes revisited,” Nat. Rev. Clin. Oncol., vol. 14, no. 10, p. 587, Oct. 2017.
B. J. Gill et al., “MRI-localized biopsies reveal subtype-specific differences in molecular and cellular composition at the margins of glioblastoma,” Proceedings of the National Academy of Sciences, vol. 111, no. 34. pp. 12550–12555, 2014. doi: 10.1073/pnas.1405839111.
“NF1 gene.” https://medlineplus.gov/genetics/gene/nf1/ (accessed Jan. 02, 2022).
“PDGFRA gene.” https://medlineplus.gov/genetics/gene/pdgfra/ (accessed Jan. 02, 2022).
“MGMT Promoter Methylation.” https://www.mayocliniclabs.com/test-catalog/overview/36733#Clinical-and-Interpretive (accessed Jan. 02, 2022).
F. S. Saadeh, R. Mahfouz, and H. I. Assi, “EGFR as a clinical marker in glioblastomas and other gliomas,” Int. J. Biol. Markers, vol. 33, no. 1, pp. 22–32, Jan. 2018.
“EGFR Gene.” https://medlineplus.gov/genetics/gene/egfr/ (accessed Jan. 02, 2022).
J. Hobbs et al., “Paradoxical relationship between the degree of EGFR amplification and outcome in glioblastomas,” Am. J. Surg. Pathol., vol. 36, no. 8, pp. 1186–1193, Aug. 2012.
M. Perkuhn et al., “Clinical Evaluation of a Multiparametric Deep Learning Model for Glioblastoma Segmentation Using Heterogeneous Magnetic Resonance Imaging Data From Clinical Routine,” Investigative Radiology, vol. 53, no. 11. pp. 647–654, 2018. doi: 10.1097/rli.0000000000000484.
A. H. Fischer, K. A. Jacobson, J. Rose, and R. Zeller, “Hematoxylin and eosin staining of tissue and cell sections,” CSH Protoc., vol. 2008, p. db.prot4986, May 2008.
Allen Institute for Brain Science, “Ivy Glioblastoma Atlas Project.” May 2015. [Online]. Available: https://glioblastoma.alleninstitute.org/
O. Ronneberger, P. Fischer, and T. Brox, “U-Net: Convolutional Networks for Biomedical Image Segmentation,” in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015, 2015, pp. 234–241.
M. Loukadakis, J. Cano, and M. O’Boyle, “Accelerating Deep Neural Networks on Low Power Heterogeneous Architectures,” presented at the Eleventh International Workshop on Programmability and Architectures for Heterogeneous Multicores (MULTIPROG-2018), Manchester, UK, Jan. 2018. Accessed: Dec. 31, 2021. [Online]. Available: https://eprints.gla.ac.uk/183819/
A. Z. Shirazi et al., “A deep convolutional neural network for segmentation of whole-slide pathology images identifies novel tumour cell-perivascular niche interactions that are associated with poor survival in glioblastoma,” British Journal of Cancer, vol. 125, no. 3. pp. 337–350, 2021. doi: 10.1038/s41416-021-01394-x.
R. Su, X. Liu, Q. Jin, X. Liu, and L. Wei, “Identification of glioblastoma molecular subtype and prognosis based on deep MRI features,” Knowledge-Based Systems, vol. 232, p. 107490, Nov. 2021.
L. Han and M. R. Kamdar, “MRI to MGMT: predicting methylation status in glioblastoma patients using convolutional recurrent neural networks,” in Biocomputing 2018, WORLD SCIENTIFIC, 2017, pp. 331–342.
I. Levner, S. Drabycz, G. Roldan, P. De Robles, J. G. Cairncross, and R. Mitchell, “Predicting MGMT Methylation Status of Glioblastomas from MRI Texture,” in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2009, 2009, pp. 522–530.
Y. Li et al., “MRI features can predict EGFR expression in lower grade gliomas: A voxel-based radiomic analysis,” Eur. Radiol., vol. 28, no. 1, pp. 356–362, Jan. 2018.
Additional Project Information
Research Plan:
Rationale
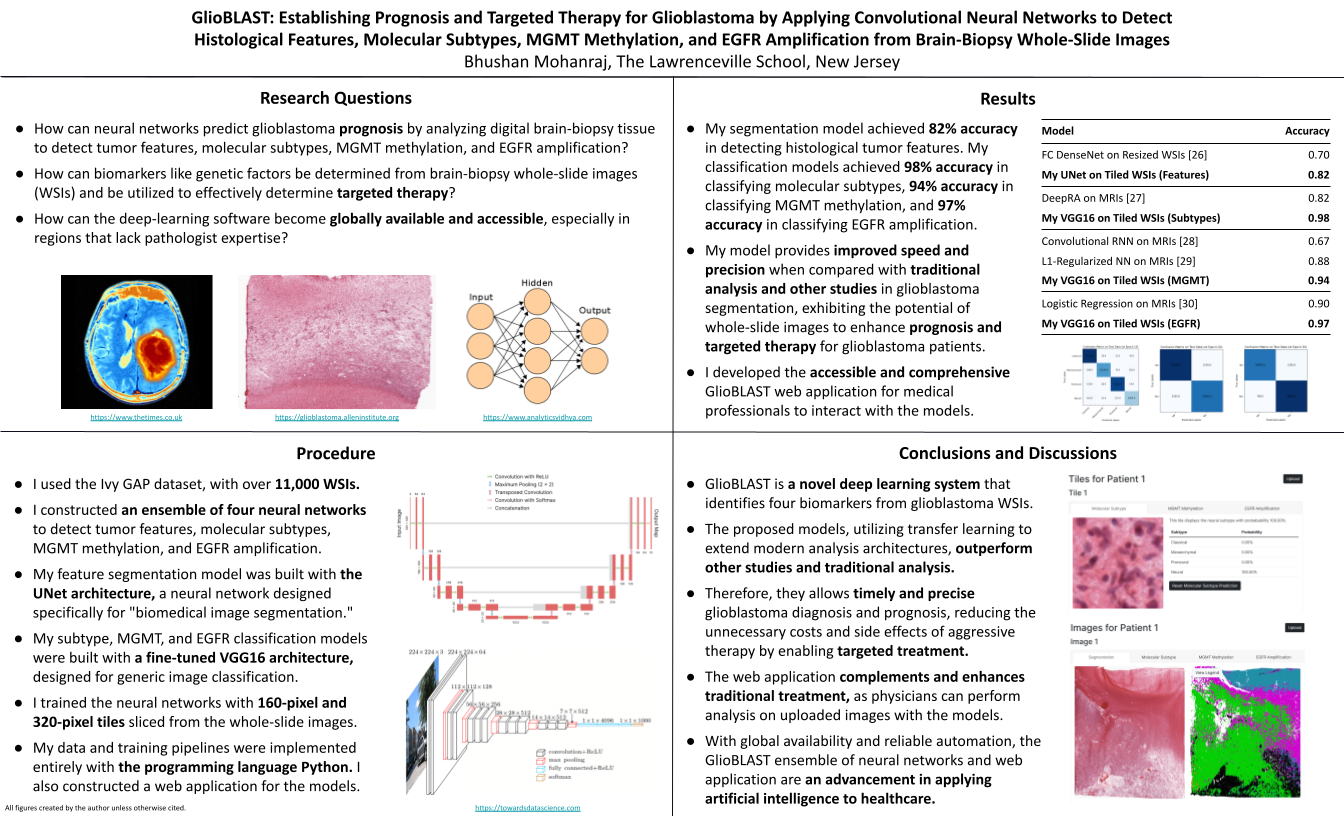
Glioblastoma is the most aggressive and deadly brain cancer, having a survival rate of 7% after five years and accounting for 48% of all primary malignant brain tumors. However, early detection and classification can significantly improve patient prognosis. Diagnosis and prognosis rely extensively on identifying tumor features (like pseudopalisading necrosis) and determining biomarkers and genetic factors (like MGMT gene methylation and EGFR gene amplification) from biopsy tissue and imagery. However, manual examination requires days for diagnosis and encounters variability between pathologists. Furthermore, many areas around the world lack pathologists with the specific expertise needed for diagnoses. Recent advances in cancer care have focused heavily on therapy targeting specific genetic characteristics of disease, and this approach could also apply to glioblastoma. Deep learning and computer vision can address these constraints with accurate image analysis and targeted therapy. I plan to employ the UNet and VGG16 neural-network architectures to perform segmentation of tumor features and classification for molecular subtypes, MGMT methylation, and EGFR amplification from whole-slide images. I also plan to create a web application through which medical professionals can interact with my computational models.
Research Questions
- How can neural networks predict glioblastoma prognosis by analyzing digital brain-biopsy tissue to detect tumor features, molecular subtypes, MGMT methylation, and EGFR amplification?
- How can biomarkers like genetic factors be determined from brain-biopsy whole-slide images and be utilized to effectively determine targeted therapy?
- How can the deep-learning software become globally available and accessible, especially in regions that lack pathologist expertise?
Goal
To develop a novel deep learning application for precisely predicting glioblastoma prognosis with less time and greater accuracy than conventional methods by utilizing artificial intelligence in biomedical image analysis.
Procedure
- Analyze the hardware requirements that are needed for handling digital whole-slide images and training the neural-network model.
- Set up a workspace with the necessary memory and software packages to train the models with the laptop GPU.
- Explore the REST API provided by Ivy GAP project to obtain the whole-slide images, ground-truth masks, and tumor-feature metadata.
- Perform exploratory analysis, and consider the sizes of the images before data processing.
- Identify data-preprocessing methods such as patching, tiling, color stabilization, and normalization. Identify data-augmentation methods such as flipping and cropping.
- Compare manual feature extraction using machine-learning algorithms (including SVMs and XGBoost) and automatic extraction using deep-learning algorithms (including CNNs). Explore transfer learning and fine tuning for pretrained models such as the VGG16 and ResNet architectures.
- Implement custom TensorFlow callbacks to store model checkpoints, create metric plots, and produce the confusion matrix after each training epoch.
- Analyze and implement custom losses such as weighted categorical cross-entropy, the Dice coefficient, and the generalized Dice coefficient.
- Create model prototypes and four training pipelines for the models. Optimize hyperparameters such as the learning rate, batch size, and loss. Tune the hyperparameters and save the optimal model based on the accuracy and loss metrics.
- Consider different frameworks for developing the web application, focusing one the flexibility and extensibility of each.
- Build the frontend and backend of the web application with the model-view-controller architecture, using Python frameworks such as Django, Flask, SQLAlchemy, and WTForms.
- Load the saved models into the web application, and allow users to receive predictions after uploading their own whole-slide images.
- Sign up for Amazon Web Services or PythonAnywhere to configure the web application. After testing, deploy the web application publicly for general use.
Data Analysis
- Use custom TensorFlow callbacks to plot the accuracy and loss metrics and the confusion matrix after each training epoch.
- Optimize the hyperparameters and loss functions by analyzing the metric plots after attempting different training configurations. Training should be repeated as needed.
Loss Functions
Segmentation Categorical Cross-Entropy Loss Function
The categorical cross-entropy loss function for training the segmentation model is
where T and O are the target tensor (from the dataset) and output tensor (from the model), respectively. The tensors have shapes (I,R,C,N), corresponding to I batch images, R rows and C columns, and N classes. The element for image i at row r and column c represents the probability distribution of the classes for the corresponding image pixel.
Classification Categorical Cross-Entropy Loss Function
The categorical cross-entropy loss function for training the classification models is
where T and O are the target tensor (from the dataset) and output tensor (from the model), respectively. The tensors have shapes (I,R,C,N), corresponding to I batch images, R rows and C columns, and N classes. The element for image i at row r and column c represents the probability distribution of the classes for the corresponding image pixel.
Classification Binary Cross-Entropy Loss Function
The binary cross-entropy loss function for training the classification models is
where T and O are the target tensor (from the dataset) and output tensor (from the model), respectively. The tensors have shapes (I), corresponding to I batch images. The element for image i represents the probability that the image exhibits the property for the model.
Performance Metrics
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
TP = True Positives; TN = True Negatives; FN = False Negatives; FP = False Positives
Risk and Safety
There are no risks associated with this project.
Questions and Answers
1. What was the major objective of your project and what was your plan to achieve it?
Through my project, I intended to construct deep-learning models to determine prognosis and targeted therapy for glioblastoma, a malignant brain cancer, from brain-biopsy whole-slide images to improve the speed and accuracy of patient treatment. Furthermore, I intended to make these models publicly accessible through a web application, enabling global access to expert-level analysis, especially within regions that lack pathologist expertise.
To achieve this objective, I considered and researched multiple data modalities for glioblastoma, including magnetic-resonance images (MRIs), computed tomography scans (CT scans), and whole-slide images (WSIs). To produce neural networks for enhancing pathologist analysis, I explored biomarkers that pathologists typically analyze when determining tumor progression and targeted treatment options. These biomarkers include tumor features, molecular subtypes, MGMT methylation, and EGFR amplification.
I chose the extensive Ivy GAP dataset for conducting my project. Ivy GAP provides over eleven-thousand whole-slide images, more than any other glioblastoma collection, and provides values of these four biomarkers for every whole-slide image. Using the dataset, I planned to extract tiles from the whole-slide images and to train computer-vision models to determine biomarkers for prognosis and targeted treatment. After building these models, I planned to construct a public web application to enable medical professionals to upload their whole-slide images and receive predictions produced by the application running the models.
a. Was that goal the result of any specific situation, experience, or problem you encountered?
I have engaged in scientific research for many years, and my recent projects have propelled my interest in programming and computational biology. Although I programmed for many years before, I specifically entered the domain of deep learning last year, when I implemented my first neural networks with Python and began appreciating the substantial potential of deep learning to improve lives.
My uncle was diagnosed with oral cancer three years ago and underwent surgery after his tumor progressed significantly. The experience of managing and responding to a family cancer diagnosis propelled me to apply my interests and knowledge in programming and deep learning to oncology. While researching cancers, I encountered glioblastoma, the most aggressive brain cancer from which almost all patients die. However, doctors minimally understand the disease and its inevitable recurrence. I thus decided to explore how neural networks could improve initial prognoses and guide targeted therapies for glioblastoma.
b. Were you trying to solve a problem, answer a question, or test a hypothesis?
I intended to solve the slow diagnosis of glioblastoma and the variations between pathologists when classifying tumors. After an initial resection surgery, pathologists manually examine tumor tissue to determine histological features and genetic factors influencing tumor progression. However, the manual examination of tissue requires up to ten days, a crucial period for responding to aggressive tumors. Tissue examination also encounters variability between pathologists, preventing patients from receiving essential targeted therapy.
I also intended to determine whether deep learning could effectively provide prognostic systems and achieve precision and clinical viability in analyzing glioblastoma. I specifically explored whole-slide images and their potential, which few studies have explored when implementing deep learning for glioblastoma.
2. What were the major tasks you had to perform in order to complete your project?
When beginning my project, I considered various data sources to determine which could provide extensive and specific data for glioblastoma tumors. I eventually settled upon the Ivy GAP dataset managed by the Allen Institute for Brain Science. The Ivy GAP dataset publicly provides over eleven-thousand whole-slide images. Furthermore, the dataset provides extensive data for critical tumor biomarkers, including histological features, molecular subtypes, and genetic factors such as MGMT methylation and EGFR amplification.
After selecting my project dataset, I determined which biomarkers I could effectively employ for constructing my deep learning models. I thus chose to create a segmentation model for detecting histological features and three classification models for detecting molecular subtypes, MGMT methylation, and EGFR amplification. I then explored potential architectures for constructing precise neural networks, and I eventually chose the modern UNet architecture for segmentation and VGG16 architecture for classification.
Next, I explored potential technologies for building my segmentation and classification models. With the extensive selection of Python libraries for neural networks and website development, I selected Python as my primary programming language. After exploring frameworks for deep learning such as TensorFlow and PyTorch, I chose TensorFlow to implement my neural networks for its intuitive application programming interface (API). After exploring frameworks for website development such as Flask and Django, I chose Flask to develop my web application for its flexible routing and application structure.
After determining the datasets, architectures, and technologies for my neural networks, I began preprocessing my data for training my models. Due to the large size of each whole-slide image, with over ten thousand pixels along each axis, I tiled the whole-slide images before creating models using the data. For the segmentation model, I used 320-pixel tiles (from both the images and segmentation masks), while for the three classification models, I used 160-pixel tiles (from the images). I then constructed the neural networks with the Keras API using the UNet and VGG16 architectures.
After collecting my preprocessed data and initially constructing my models, I trained the neural networks. I trained my segmentation model with image tiles and the corresponding mask tiles, and I trained my three classification models with image tiles and the corresponding labels (such as "Classical" or "Mesenchymal" for the molecular subtype). For each model, I used 80% of my data for training and the remaining 20% for validation, and I trained the models with the standard categorical-cross-entropy loss function.
After training my models, I achieved 82% accuracy for segmenting histological features and 98%, 94%, and 97% accuracy for classifying subtypes, MGMT methylation, and EGFR amplification. My neural networks thus demonstrate clinical viability and could enhance traditional pathologist analysis. With my models, I then began designing and implementing my web application. I constructed an interface through which medical professionals can upload whole-slide images and receive predictions from the segmentation and classification models. Through Amazon Web Services, my web application is globally accessible, allowing medical professionals globally to swiftly and precisely determine glioblastoma prognosis and targeted therapy.
a. For teams, describe what each member worked on.
I worked individually on my project.
3. What is new or novel about your project?
To analyze tumor biomarkers, I employed whole-slide images, an unused data modality in computational systems for glioblastoma. From my literature review, I found numerous studies applying neural networks to MRI and CT scans but only one to whole-slide images. However, histopathological analysis of whole-slide images remains the gold standard for glioblastoma diagnosis and prognosis.
Whole-slide images contain over ten-thousand pixels along each axis, making them unwieldy in data processing when compared with other sources. Nevertheless, the data within each enables insightful analysis, as demonstrated by the accuracy of my neural networks. Computational systems using whole-slide images often resize them, thus losing minute details of structures like cells and blood vessels. I instead employed a tiling system to manage these images, where I sliced the images into smaller tiles before training my models.
Furthermore, my project includes a web application for deploying my models. Although data scientists employ web applications extensively for creating visualizations, I found no other glioblastoma studies with a web application for deploying neural networks or other computational systems. Through the web application, my GlioBLAST neural networks become accessible to medical professionals globally, especially within regions without pathologists who have the specific expertise needed for glioblastoma diagnosis.
a. Is there some aspect of your project's objective, or how you achieved it that you haven't done before?
Although I have constructed neural networks before, I have never interacted with datasets as large as Ivy GAP, which contained eleven-thousand whole-slide images, each with ten thousand pixels along each axis. Furthermore, I have never constructed an ensemble of multiple neural networks which reveal data by manipulating the same data source. Exploring these data sources and building multiple neural networks has broadened my understanding of neural networks and computational biology and has revealed the tremendous potential of deep learning.
b. Is your project's objective, or the way you implemented it, different from anything you have seen?
During my literature review, I found no studies which implement neural networks for multiple tumor biomarkers. However, by analyzing four biomarkers, I demonstrated that neural networks can reveal detailed histological and genetic information for glioblastoma tumors, improving prognosis and targeted treatment for patients. Furthermore, although I found many studies using MRI images or CT scans, I found only one other that used whole-slide images constructing computational models for glioblastoma. I also employed a novel tiling method, allowing me to efficiently manage the data within each whole-slide image.
c. If you believe your work to be unique in some way, what research have you done to confirm that it is?
I conducted an extensive literature review into studies that apply neural networks to medical challenges and, specifically, to glioblastoma biomarkers. Although neural networks have gained popularity, I found relatively little research employing deep learning for glioblastoma prognosis, especially with whole-slide images for prognosis and targeted treatment. Furthermore, I found no other studies which constructed an ensemble of neural networks for distinguishing multiple tumor biomarkers in a unified computational system. Finally, I found no other studies which deployed such a system through a web application.
4. What was the most challenging part of completing your project?
The most challenging section of completing my research occurred while preprocessing the whole-slide images from the Ivy GAP dataset. The data contained within each image presented time and memory constraints for processing. Although I initially attempted to manage the images by resizing them, my models had difficulty accurately classifying the tumors according to genetic factors since I had removed the minute details of cells and other structures. I then considered multiple methods for persevering these details, and I eventually tiled the whole-slide images into smaller sections. Tiling, however, created thousands of tiles per image, which required days to complete and prevented me from tiling the entire dataset. But after I employed the tiling approach, my models performed well across all four biomarkers.
a. What problems did you encounter, and how did you overcome them?
While conducting my research, initial challenges arose while collecting data, since the whole-slide images required unexpectedly-large computer memory. I overcame these memory challenges by downloading a subset rather than the entire dataset (which would require terabytes of data). Furthermore, I used an Amazon Web Services S3 instance for training my histological segmentation model with more CPU and GPU memory.
I later encountered challenges while tuning the neural-network hyperparameters. The segmentation model, which outputs masks with ten classes, proved especially difficult to tune. I attempted to implement custom loss functions, including weighted categorical cross-entropy, the Dice coefficient, and the generalized Dice coefficient. However, I returned to categorical cross-entropy after these functions proved less successful. I later decreased the learning rate, which reduced overfitting but lengthened training time. I finally reduced the batch size to one (the same batch size employed by the original UNet authors), enabling the segmentation model to generalize well without time or memory constraints.
b. What did you learn from overcoming these problems?
By overcoming the immense sizes of my whole-slide images, I learned to manage large datasets, which enable detailed analysis but present memory constraints for programming. By tuning the segmentation and classification models, I learned the influence of different hyperparameters (including the batch size, learning rate, loss function, and network architecture) on the performance of neural networks. To implement my loss functions, I researched the underlying mathematics behind deep learning, understood the training loop behind neural networks, and explored the comprehensive APIs of the TensorFlow and Keras frameworks.
5. If you were going to do this project again, are there any things you would you do differently the next time?
I would begin data exploration and preprocessing early since using and understanding large datasets requires patience and time. To improve training and model performance, I would invest early in an Amazon Web Services instance to complete almost all data processing and model training, rather than performing such operations through my laptop. I would finally conduct more research into neural-network architectures for constructing my models. Although I initially built custom neural networks for image classification, I later employed the VGG16 architecture with transfer learning, vastly improving training time and model performance.
6. Did working on this project give you any ideas for other projects?
Through my project, I explored neural networks and deep learning in computational biology. I have thus realized the tremendous potential of artificial intelligence to transform medicine and other fields. After constructing an ensemble of neural networks that analyzes image data, I hope to extend my computational system with more data modalities. For example, the Ivy GAP dataset includes data for genetic expression within tumors, which complement the other tumor biomarkers my models currently analyze. Furthermore, I could extend my analysis system and architectures to prognosis and targeted treatment for other tumors and diseases.
7. How did COVID-19 affect the completion of your project?
Due to the COVID-19 pandemic, I conducted my research individually since most opportunities with university mentors and laboratories were limited. Without mentorship, I had difficulty accessing the Ivy GAP dataset and downloading the available whole-slide images. Furthermore, I experienced challenges performing data preprocessing and training my models without large memory or powerful GPUs. Under future mentorship with university mentors, I would like to explore the clinical viability of my computational system and achieve enhanced performance by conducting future projects with better computational resources.