Role of Cancer Associated Fibroblast Heterogeneity on Immunotherapuetic Potentials of Pancreatic Ductal Adenocarcinoma
Abstract:
Bibliography/Citations:
[1] R. L. Siegel, K. D. Miller, and A. Jemal, “Cancer statistics, 2020,” CA: A Cancer Journal
for Clinicians, vol. 70, no. 1, pp. 7–30, 2020, doi: 10.3322/caac.21590.
[2] L. Rahib, B. D. Smith, R. Aizenberg, A. B. Rosenzweig, J. M. Fleshman, and L. M.
Matrisian, “Projecting Cancer Incidence and Deaths to 2030: The Unexpected Burden of
Thyroid, Liver, and Pancreas Cancers in the United States,” Cancer Res, vol. 74, no. 11,
pp. 2913–2921, Jun. 2014, doi: 10.1158/0008-5472.CAN-14-0155.
[3] S. Gillen, T. Schuster, C. Meyer zum Büschenfelde, H. Friess, and J. Kleeff,
“Preoperative/Neoadjuvant Therapy in Pancreatic Cancer: A Systematic Review and
Meta-analysis of Response and Resection Percentages,” PLoS Med, vol. 7, no. 4, p.
e1000267, Apr. 2010, doi: 10.1371/journal.pmed.1000267.
[4] Y. Wang et al., “Single-cell analysis of pancreatic ductal adenocarcinoma identifies a
novel fibroblast subtype associated with poor prognosis but better immunotherapy
response,” Cell Discov, vol. 7, no. 1, pp. 1–17, May 2021, doi:
10.1038/s41421-021-00271-4.
[5] Y. Kimura et al., “Clinical and immunologic evaluation of dendritic cell-based
immunotherapy in combination with gemcitabine and/or S-1 in patients with advanced
pancreatic carcinoma,” Pancreas, vol. 41, no. 2, pp. 195–205, Mar. 2012, doi:
10.1097/MPA.0b013e31822398c6.
[6] M. Reck et al., “Five-Year Outcomes With Pembrolizumab Versus Chemotherapy for
Metastatic Non–Small-Cell Lung Cancer With PD-L1 Tumor Proportion Score ≥ 50%,”
JCO, vol. 39, no. 21, pp. 2339–2349, Jul. 2021, doi: 10.1200/JCO.21.00174.
[7] M. Orth et al., “Pancreatic ductal adenocarcinoma: biological hallmarks, current status,
and future perspectives of combined modality treatment approaches,” Radiation
Oncology, vol. 14, no. 1, p. 141, Aug. 2019, doi: 10.1186/s13014-019-1345-6.
[8] L. Monteran and N. Erez, “The Dark Side of Fibroblasts: Cancer-Associated Fibroblasts
as Mediators of Immunosuppression in the Tumor Microenvironment,” Frontiers in
Immunology, vol. 10, p. 1835, 2019, doi: 10.3389/fimmu.2019.01835.
[9] Y. Binenbaum, S. Na’ara, and Z. Gil, “Gemcitabine resistance in pancreatic ductal
adenocarcinoma,” Drug Resist Updat, vol. 23, pp. 55–68, Nov. 2015, doi:
10.1016/j.drup.2015.10.002.
[10] H. Jiang et al., “Targeting focal adhesion kinase renders pancreatic cancers responsive to
checkpoint immunotherapy,” Nat Med, vol. 22, no. 8, pp. 851–860, Aug. 2016, doi:
10.1038/nm.4123.
Additional Project Information
Research Plan:
- Rationale
Pancreatic Ductal Adenocarcinoma (PDAC) has the poorest prognosis of all cancers with a mere 9% 5-year survival rate. Pancreatic cancer kills close to 50,000 people each year making it the 4th leading cause of cancer death worldwide. One of the main challenges with PDAC is the minimal responses to treatment with only a 20% chemotherapy success rate and a mere 1-2% approval for PDAC immunotherapies. A major contributor to the limited treatment successes is the dense stroma or fibrosis that surrounds the tumor. The fibrosis, which makes up to 90% of tumor volume, creates a biophysical barrier around the tumor preventing drug penetration. This excess fibrosis stems from cancer-associated fibroblasts (CAFs) which are a subpopulation of cells that regulate the tumor microenvironment of PDAC. Targeting CAFs may allow for better treatment outcomes, however, due to the high heterogeneity of CAFs there is currently a poor understanding of the stromal-immune crosstalk defined as the relationship between fibroblasts and tumor immune cells that affects treatment response. Only by understanding the differential effect of CAF subpopulations on the immune microenvironment of PDAC can pro-tumoral subtypes be targeted and more effective immunotherapies be created. The purpose of this data analysis study is to understand CAF functional heterogeneity in immune cell regulation to help identify subsets of patients that could respond favorably to immunotherapies, thereby allowing for more personalized, effective treatment.
- Research Questions, Hypotheses, Goals, Expected Outcomes
- Research Questions
- What is the role of cancer-associated fibroblast heterogeneity on the tumor microenvironment of PDAC?
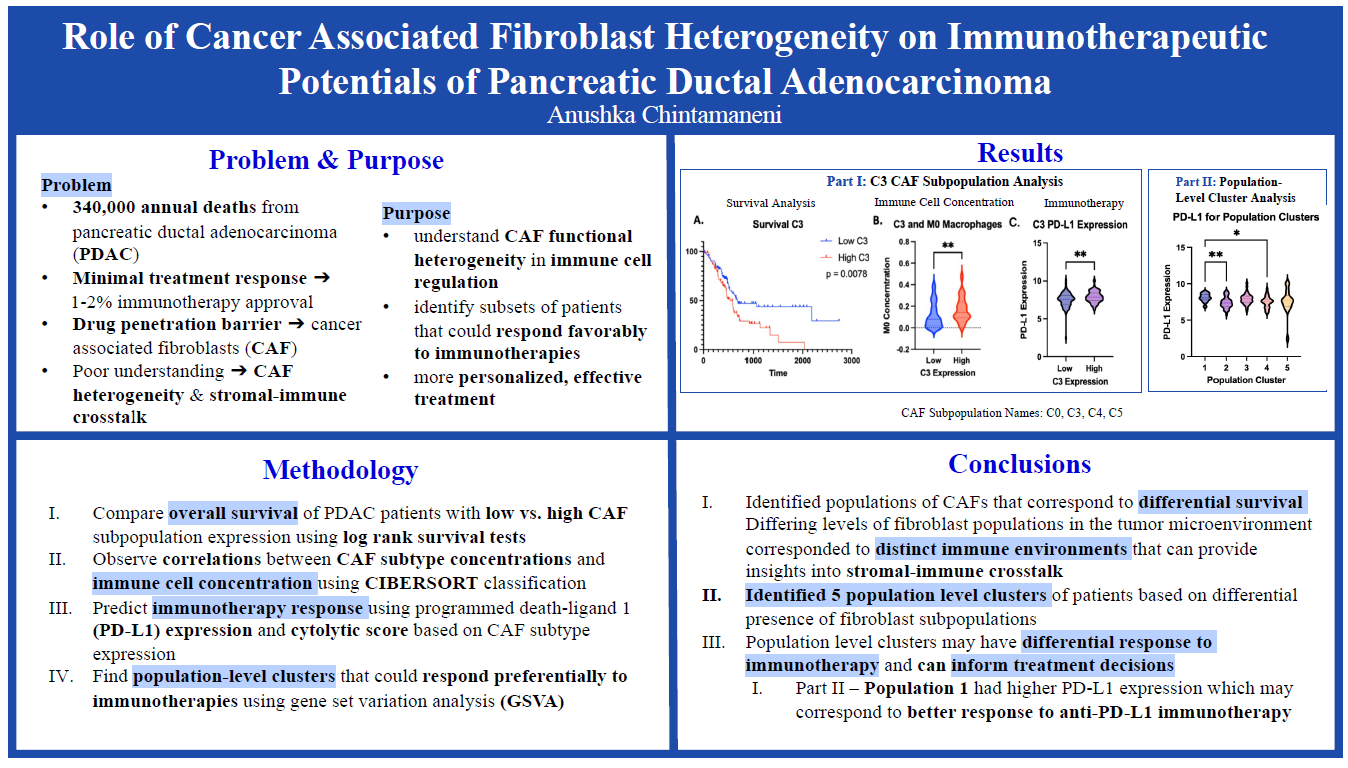
- Does CAF subpopulation concentration have an effect on the survival outcomes and immune cell concentrations of patients with PDAC?
- Is the presence of CAFs associated with poorer immunotherapeutic outcomes? Does targeting CAFs lead to better immunotherapeutic outcomes?
- Hypotheses
- If patients have higher concentrations of a CAF subpopulation, the patients will have poorer survival outcomes.
- If there are fewer CAFs, immunotherapies will have greater efficacy, thereby leading to better overall survival.
- Different CAF subpopulations will have differing effects on the immunotherapeutic outcomes of patients with PDAC.
- Research Questions
- Procedure and Data Analysis
- PDAC bulk RNA-seq dataset obtained from The Cancer Genome Atlas (TCGA, available online: https://cancergenome.nih.gov/) will be used. Marker genes CAF subpopulations will be obtained from Supplementary Table S6 assembled by Wang et al. Data normalization and tests will be performed using R in RStudio.
- Log-rank survival tests will be performed in Prism Graphpad to determine the differential survival between patients with high-expressions and low-expressions of CAF subpopulations.
- Relative immune cell concentrations in PDAC patients will be calculated using Cell-type Identification by Estimating Relative Subsets of RNA Transcripts (CIBERSORT).
- Results of CIBERSORT will be analyzed and plotted in Prism Graphpad using multiple t-tests and one-way ANOVA analysis.
- Gene Set Variation Analysis (GSVA) will be used to cluster patients based on the differential expression of multiple CAF subpopulations.
- The pheatmap library in R will be used to delineate and graphically interpret clusters found through GSVA.
- Cytolytic score (CYT score) which correlates with CD8+ T cell infiltration will be calculated by taking the geometric mean of mRNA expression levels of granzyme (GZMA) and perforin (PRF1) in the clusters.
- Results of the CYT score will be analyzed and plotted in Prism Graphpad using multiple t-tests and one-way ANOVA analysis.
- Programmed Death-Ligand 1(PD-L1) Expression, a predictive biomarker to determine the efficacy of immune checkpoint blockade, will be organized using Microsoft Excel in order to determine the predicted efficacy of PD-L1 immunotherapy based on CAF expression.
- Results of PD-L1 expression will be analyzed and plotted in Prism Graphpad using multiple t-tests and one-way ANOVA analysis.
- A p-value of less than 0.05 will be considered significant for all statistical tests performed.
- Risk and Safety
- The study poses no risk to safety as all experimentation will be done computationally.
Questions and Answers
1. What was the major objective of your project and what was your plan to achieve it?
One of the main challenges with cancer treatment is the complexity and uniqueness of each tumor. When I was younger, I used to think that there could be one singular cure for cancer. Sadly, cancer isn’t as linear as I had hoped. Tumors each have their own unique microenvironment with hundreds of different signaling pathways, growth factors, and immune cells that factor into their aggressiveness. Because of the complex nature of tumors, there’s no ‘one size fits all’ for cancer treatment. Therefore, in order to combat the problem of generalized treatment programs which may not address the specific characteristics of a person's tumor, I wanted to look into ways to personalize treatment options in hopes of improving efficacy and survival outcomes. I wanted to do a bioinformatics-based project because of the extensive data analytics capabilities and access to datasets containing comprehensive information of an abundance of tumors.
a. Was that goal the result of any specific situation, experience, or problem you encountered?
A few years ago, my friend’s mom got a cancer diagnosis and I saw the devastating effects on her family as a whole. I can only begin to imagine the pain associated with chemotherapy treatments and the fear that accompanies a cancer diagnosis. I hated just sitting idly by knowing that people were suffering every day from such a devastating condition and I wanted to take action. I took my passion for medicine and applied it to a real-world problem.
b. Were you trying to solve a problem, answer a question, or test a hypothesis?
I specifically chose to focus on pancreatic ductal adenocarcinoma (PDAC) because it has the poorest prognosis of all cancer types with a mere 9% 5-year survival rate. I wanted to answer the question of what contributes to the aggressiveness of PDAC and solve the problem of ineffective treatment by looking into more personalized options. I focused on immunotherapies because I wanted to see how the complex immune microenvironment of PDAC affected immunotherapeutic treatment.
2. What were the major tasks you had to perform in order to complete your project?
First and foremost, in order to complete my project, I had to do a comprehensive literature review. I wanted to make sure my research was novel and not repetitive so I read a lot of cancer journals to find ways in which I could effectively contribute to current findings. After deciding a topic, I worked on outlining the major parts of my project by writing my research plan. I detailed different bioinformatics techniques that I had researched and thought I could use. With help from my mentor, I normalized the PDAC dataset and conducted all of my tests using RStudio, corresponding packages, Excel, and PRISM Graphpad. After completing the data analytical portion of my research, I got to work on contextualizing my results. Getting positive correlations between cell types has little meaning unless applied to a certain context. I did thorough research to interpret what my results meant and why it was important. I then started to pull together all of my research and wrote a research paper and made a poster presentation.
3. What is new or novel about your project?
My project specifically addressed the poorly understood area of cancer associated fibroblasts (CAFs). Because CAFs are highly heterogeneous, it has been difficult to characterize the pro-tumoral and anti-tumoral functions as well as the stromal-immune crosstalk (how fibrosis affects the immune environment) of a tumor. I specifically identified five new population-level clusters based on a combination of CAF subtypes and analyzed immunotherapeutic markers in each population to determine which patients could respond better to anti-PD-L1 immunotherapy. Based on this information, more personalized treatment options can be formed that improve efficacy and a greater understanding of CAF heterogeneity and target opportunities is achieved.
a. Is there some aspect of your project's objective, or how you achieved it that you haven't done before?
Prior to working on this project, I had only ever read bioinformatics papers and cancer journals. It was a very unique experience to conduct independent bioinformatics research and apply some of the concepts and techniques I had only ever read about. It’s fascinating to see how something as small as one specific cell type can have a major impact on a whole system or disease through complex connections.
b. If you believe your work to be unique in some way, what research have you done to confirm that it is? Is your project's objective, or the way you implemented it, different from anything you have seen?
As I thought about my project topic, I wanted to contribute something useful and new to the scientific community. For this reason, I made sure that the research I was conducting was not repeating someone else's work. I poured over hundreds of academic journals in order to familiarize myself with all things cancer-related. I learned all about the immune microenvironment of PDAC that affected survival. I found that although CAFs are a major part of PDAC aggressiveness, the heterogeneity of this cell type negatively impacted a lot of the present research. There needed to be a better understanding of CAF classifications in order to target the pro-tumoral subtypes and amplify the effects of anti-tumoral subtypes. Therefore, I focused on a unique connection between CAFs and the immune landscape of PDAC as well as the immunotherapeutic potentials. As I stated above, I conducted a thorough analysis of CAFs that both provide greater insights into how to target this cell type but also allow for more personalized therapy options based on the five population-level clusters identified.
4. What was the most challenging part of completing your project? What problems did you encounter, and how did you overcome them? What did you learn from overcoming these problems?
Some of the challenges I ran into were data normalization errors and data interpretation. When I first collected my dataset and attempted to normalize it, I ran into quite a few errors. In order to fix this problem, I troubleshooted in many different ways such as redownloading my dataset, using another dataset, and researching common problems with the Biocmanager package I was using. Eventually, I was able to correctly normalize my data and start conducting all of my tests. I applied the same troubleshooting methods when I ran into problems with my Gene Set Variation Analysis code. Additionally, after organizing my data and inputting it into PRISM GraphPad, I had to decide which graphs and tests best compared my data to make the most meaningful conclusions. I decided upon one-way ANOVA tests using violin plots as it allowed for multiple comparisons and graphically showed data distribution.
5. If you were going to do this project again, are there any things you would do differently the next time?
For the most part, the computational aspect of my project went well. However, if I were to do this project again, during the literature review, I would take the time to organize all of the articles I have read. After a while, I got so engrossed in the content of what I was reading that I would forget to organize each journal into different categories. Instead of a master file of hundreds of articles, if each article is put into separate categories, it could lead to easier access and better organization. For example, if I wanted to recall a specific statistic relating to angiogenesis in PDAC, I would have a separate section for ‘angiogenesis’ instead of combing through hundreds of pages of notes.
6. Did working on this project give you any ideas for other projects?
Yes, this research opened up many opportunities for further analysis and other projects, both bioinformatics-based and wet lab based. The same bioinformatics approaches used in this project can be used for other cancer types. For example, contrasting the effect of cancer associated fibroblasts across cancer types. Additionally, I can use techniques such as Gene Set variation Analysis (GSVA) and Cell-Type Identification By Estimating Relative Subsets of RNA Transcripts (CIBERSORT) to determine the connection between different signaling pathways and immune cell concentrations in a tumor microenvironment. As an extension of the presented project, further in vitro analyses can be conducted.
7. How did COVID-19 affect the completion of your project?
Because my project was bioinformatics based and conducted at home using publicly available data, COVID-19 did not affect the completion of my research.